CPC Web Server Quick Guide
CPC Web Server Quick Guide

-
Introduction
-
Coding Potential Calculator distinguish protein-coding from non-coding RNAs based on the sequence features of the input transcripts. Our preliminary performance assessment suggests the CPC can reliably discriminate the coding and non-coding transcripts in ~98% accuracy. We provide an online version of CPC here.
-
CPC Online Input
-
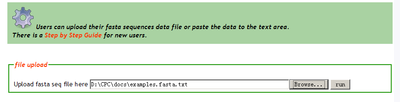
CPC online input page: Accept fasta format sequences as input. Users can upload their sequences file from local disk or just paste the sequence to the textarea of the input page. For more information about fasta format, refer to wikipeida fasta format description or ncbi fasta format description.
Example data: sequences in FASTA format .
Besides the stand FASTA format data, CPC also supports one single sequence without ID line. If user inputs an sequence in the format like THIS, CPC will assign an ID to the sequence automatically. CPC online calculator also supports mutiple sequences seperated by ">\n" . If user inputs sequences in the format like THIS, CPC will assign an ID for each sequence automatically.
-
Users can upload their local sequence file to the server.
-
Users can copy & paste their sequence data into the textarea instead of uploading the sequence file.
-
CPC is robust for low-quality transcripts, which means it is tolerant for few frameshifts or stop codons caused by sequencing errors. But, it does not intend to mean CPC is also robust for those partial transcripts, such as EST derived mostly from UTR regions. Our current core SVM classifier could not discriminate the UTR regions from non-coding transcripts, given that fact that they do not actually produce amino acid sequences. However, in most mammalian genomes, the 3' UTRs of a coding transcript can extend for several kb and are abundant in many current EST libraries. And it is often helpful to distinguish UTR regions in partial coding sequences from the standalone functional non-coding transcripts in practice. To help users to identify ESTs derived from known UTR sequences, we provided the BLAST searching against UTRdb as a supplementary module for CPC web server.
-
-
CPC Results Output
-
Users can view CPC results in its raw data format and in html format, which is more friendly.
-
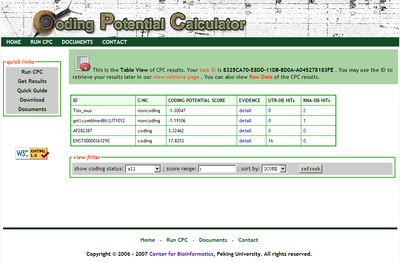
CPC resluts html view gives an overview of coding status of the input sequences. Each row corresponds to one input sequence. The columns show the sequence ID, the coding/Noncoding classification, the SVM score (the "distance" to the SVM classification hyper-plane in the features space), the "Evidence Details" link (as described later), the UTRdb search hits number and the RNAdb search hits number. For users' convenience, the results can be filtered by coding status and score, as well as be sorted by ID, coding status and score. The score filter read the score range seperated by ':' . For example, if user only wants to show the results entries whose score between -1.3 and 6.5, then the user should put "-1.3:6.5" (exclude the quot); to the score filter input area. For all entries, just left the score filter input area blank or only type a ':'.
screenshot: -
CPC online server also gives the original calculating results file. Please go to the Data Retrieve Page and use your task ID to retrieve the raw results data. The cpc raw results data file contains four columns seperated by
tab, and each line stands for the result of an input sequence.
For example:AF282387 528 coding 3.32462 Tsix_mus 4300 noncoding -1.30047 Evf1_Rat 2704 noncoding -0.991937 ENST00000361290 7834 coding 17.7115
The first column is input sequence ID; the second column is input sequence length; the third column is coding status and the four column is the coding potential score (the "distance" to the SVM classification hyper-plane in the features space).
-
-
CPC Evidence Output
-
Users can view CPC evidence features in its raw data format and in html format, which is more friendly.
-
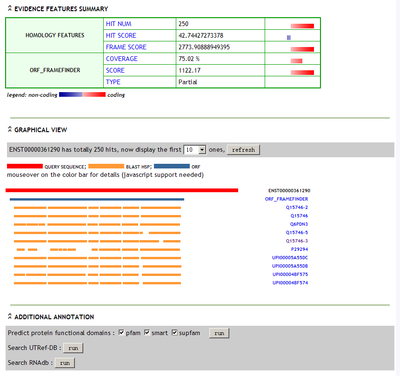
To "explain" why a transcript is classified as coding or noncoding, CPC server provides detailed supporting evidence and other related sequence features of the input transcript in an Evidence page (Figure 1b). The html view of CPC evidence features includes the following six sections :
- EVIDENCE FEATURES SUMMARY
- GRAPHICAL VIEW
- ADDITIONAL ANNOTATION
- ORF INFORMATION
- PUTATIVE PEPTIDE
- BLAST SUMMARY
 [+] view large picture
[+] view large picture
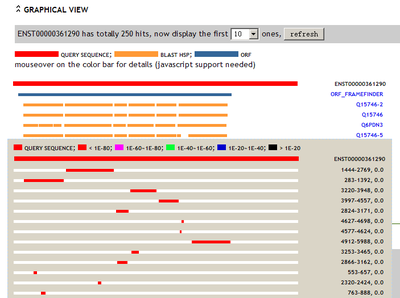
Mousing over the orf and blastx hsp color bar, users can view details of each ORF and BLASTX hits. For example:
 [+] view large picture
[+] view large picture
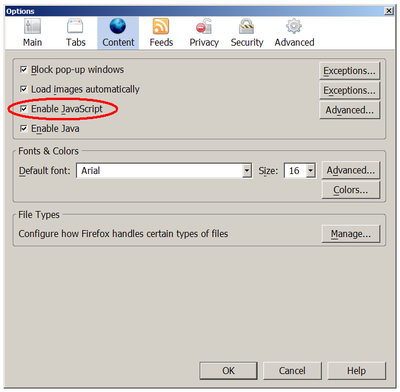
To enable this fuction, the client web browser should enable javascript support. Refer to our attachment help to see how to enable javascript support in Firefox.
-
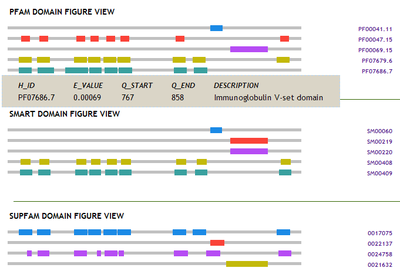
For further investigation, user can also query the putative peptide translated from ORF found by framefinder against the functional domain database Pfam, SMART and SuperFamily.
screenshot:
 [+] view large picture
[+] view large picture
Clicking the right hit ID will lead to the record of corresponding database site.
-
The evidence page can also let user query their input sequence againts UTR sequence database UTResource-DB (UTRdb).
screenshot:
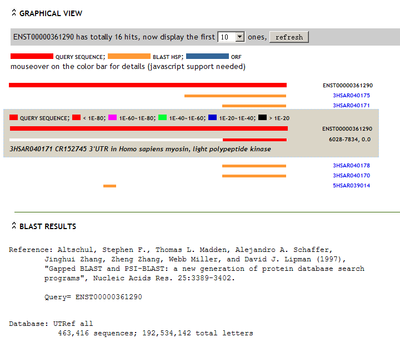 [+] view large picture
[+] view large picture
-
The evidence page can also let user query their input sequence againts non-coding RNA database RNAdb.
screenshot:
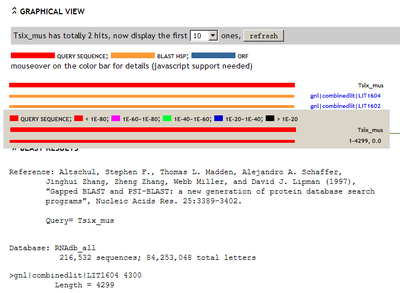 [+] view large picture
[+] view large picture
-
-
Data Retrieve Page
-
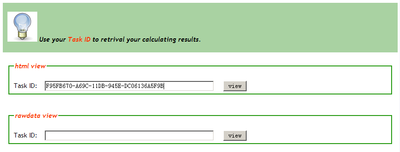
When user gives input sequences to the CPC web server for calculating, the web server will assign an unique Task ID to this request. The task id is in the format like F95FB670-A69C-11DB-945E-DC06136A5F9B . After finishing the calculating, users can go to the Data Retrieve Page and use the Task ID to retrieve their results from CPC server. For example:
-
Attachment Help
-
-
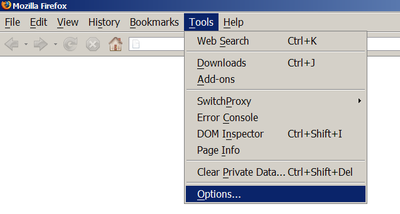
How to enable javascript in Firefox?Tools -> Options -> Content -> Enable Javascript
-