 This guide will give you a quick look at how to using CPC on line web tools.
If you want to run CPC on your computer, please refer to our
Installation Guide.
This guide will give you a quick look at how to using CPC on line web tools.
If you want to run CPC on your computer, please refer to our
Installation Guide.
-
 Fasta
Fasta
-
 "A sequence in FASTA format begins with a single-line description,
followed by lines of sequence data. The description line is distinguished
from the sequence data by a greater-than (">") symbol in the first column. (ncbi)"
For more information about fasta format, refer to
wikipeida fasta format description or
ncbi fasta format description.
"A sequence in FASTA format begins with a single-line description,
followed by lines of sequence data. The description line is distinguished
from the sequence data by a greater-than (">") symbol in the first column. (ncbi)"
For more information about fasta format, refer to
wikipeida fasta format description or
ncbi fasta format description.
-
 Email
Email
-
 Because of the complexity of coding potential calculating, it might takes several
minutes or more to finish the calculating. By providing email address here, users
can recieve their results by email for those long-time runing calculating.
The email address provided by users is only used for sending calculating results,
and we will not keep the email address or leak it to others.
Because of the complexity of coding potential calculating, it might takes several
minutes or more to finish the calculating. By providing email address here, users
can recieve their results by email for those long-time runing calculating.
The email address provided by users is only used for sending calculating results,
and we will not keep the email address or leak it to others.
-
 Task ID
Task ID
-
 The online coding potential calculator will automatically assign an unique ID for each
calculating task. Users can use the Task ID to retrieve their calculating results.
For more details, please refer to our Quick Guide .
The online coding potential calculator will automatically assign an unique ID for each
calculating task. Users can use the Task ID to retrieve their calculating results.
For more details, please refer to our Quick Guide .
 NOTES: Task ID may be expired
after 7 days when it is created. The results of an expired task will removed from
the CPC server and can no longer be retrieved.
NOTES: Task ID may be expired
after 7 days when it is created. The results of an expired task will removed from
the CPC server and can no longer be retrieved.
-
 Hit Num
Hit Num
-
 How many blast hits are found for the input sequence.
How many blast hits are found for the input sequence.
-
 Hit Score
Hit Score
-
 For a true protein-coding transcript the hits are also likely to have higher
quality; i.e., the HSPs (High-scoring Segment Pairs) overall tend to have
lower E-value. Thus we define feature HIT SCORE as follows::
For a true protein-coding transcript the hits are also likely to have higher
quality; i.e., the HSPs (High-scoring Segment Pairs) overall tend to have
lower E-value. Thus we define feature HIT SCORE as follows::
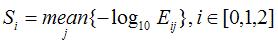
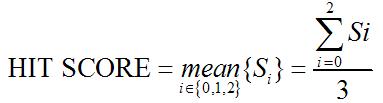
where Eij is the E-value of the jth HSP in frame i, Si measures the average
quality of the HSPs in frame i and HIT SCORE is the average of Si across three
frames. The higher the HIT SCORE, the better the overall quality of the hits
and the more likely the transcript is protein-coding.
-
 Frame Score
Frame Score
-
 For a true protein-coding transcript most of the hits are likely to reside
within one frame, whereas for a true noncoding transcript, even if it matches
certain known protein sequence segments by chance, these chance hits are
likely to scatter in any of the three frames. Thus we define
feature FRAME SCORE to measure the distribution of the HSPs
among three reading frames:
For a true protein-coding transcript most of the hits are likely to reside
within one frame, whereas for a true noncoding transcript, even if it matches
certain known protein sequence segments by chance, these chance hits are
likely to scatter in any of the three frames. Thus we define
feature FRAME SCORE to measure the distribution of the HSPs
among three reading frames:
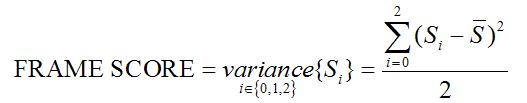
The higher the FRAME SCORE, the more concentrated the hits are and the more
likely the transcript is protein-coding.
-
 FrameFinder ORF Coverage
FrameFinder ORF Coverage
-
 FrameFinder's orf coverage, A large COVERAGE OF THE PREDICTED ORF is
an indicator of good ORF quality (Slater, G.S.C. (2000) Algorithms
for the Analysis of Expressed Sequence Tags, University of Cambridge, Cambridge.) .
For more information, refer to
Pasteur FrameFinder Man Page
.
FrameFinder's orf coverage, A large COVERAGE OF THE PREDICTED ORF is
an indicator of good ORF quality (Slater, G.S.C. (2000) Algorithms
for the Analysis of Expressed Sequence Tags, University of Cambridge, Cambridge.) .
For more information, refer to
Pasteur FrameFinder Man Page
.
-
 FrameFinder LOG-ODDS SCORE
FrameFinder LOG-ODDS SCORE
-
 As suggested by the FrameFinder's author, the LOG-ODDS SCORE is an indicator
of the quality of a predicted ORF and the higher score, the higher the quality.
For more information, refer to
Pasteur FrameFinder Man Page
.
As suggested by the FrameFinder's author, the LOG-ODDS SCORE is an indicator
of the quality of a predicted ORF and the higher score, the higher the quality.
For more information, refer to
Pasteur FrameFinder Man Page
.
-
 FrameFinder ORF Type
FrameFinder ORF Type
-
 The ORF Type is the INTEGRITY OF THE PREDICTED ORF, that indicates whether an
ORF begins with a start codon and ends with an in-frame stop codon.
The ORF Type is the INTEGRITY OF THE PREDICTED ORF, that indicates whether an
ORF begins with a start codon and ends with an in-frame stop codon.
-
 Html View
Html View
-
 The html view of coding potential calculator online results.
The html view of coding potential calculator online results.
-
 UTRdb
UTRdb
-
 UTResource-DB (UTRdb) is a specialized database of 5' and 3' UnTRanslated sequences of eukaryotic
mRNAs cleaned and annotated based on RefSeq. For more detials, refer to
UTResource-DB Web Site.
UTResource-DB (UTRdb) is a specialized database of 5' and 3' UnTRanslated sequences of eukaryotic
mRNAs cleaned and annotated based on RefSeq. For more detials, refer to
UTResource-DB Web Site.
-
 RNAdb
RNAdb
-
 A comprehensive mammalian noncoding RNA database, includes >800 unique
experimentally studied ncRNAs, >1100 putative antisense ncRNA and almost 20000
putative ncRNAs identified in high-quality murine and human cDNA libraries.
For more detials, refer to RNAdb Web Site.
A comprehensive mammalian noncoding RNA database, includes >800 unique
experimentally studied ncRNAs, >1100 putative antisense ncRNA and almost 20000
putative ncRNAs identified in high-quality murine and human cDNA libraries.
For more detials, refer to RNAdb Web Site.
 This guide will give you a quick look at how to using CPC on line web tools.
If you want to run CPC on your computer, please refer to our
Installation Guide.
This guide will give you a quick look at how to using CPC on line web tools.
If you want to run CPC on your computer, please refer to our
Installation Guide.

 This guide will give you a quick look at how to using CPC on line web tools.
If you want to run CPC on your computer, please refer to our
Installation Guide.
This guide will give you a quick look at how to using CPC on line web tools.
If you want to run CPC on your computer, please refer to our
Installation Guide.
 Fasta
Fasta
 "A sequence in FASTA format begins with a single-line description,
followed by lines of sequence data. The description line is distinguished
from the sequence data by a greater-than (">") symbol in the first column. (ncbi)"
For more information about fasta format, refer to
wikipeida fasta format description or
ncbi fasta format description.
"A sequence in FASTA format begins with a single-line description,
followed by lines of sequence data. The description line is distinguished
from the sequence data by a greater-than (">") symbol in the first column. (ncbi)"
For more information about fasta format, refer to
wikipeida fasta format description or
ncbi fasta format description.
 Email
Email
 Because of the complexity of coding potential calculating, it might takes several
minutes or more to finish the calculating. By providing email address here, users
can recieve their results by email for those long-time runing calculating.
The email address provided by users is only used for sending calculating results,
and we will not keep the email address or leak it to others.
Because of the complexity of coding potential calculating, it might takes several
minutes or more to finish the calculating. By providing email address here, users
can recieve their results by email for those long-time runing calculating.
The email address provided by users is only used for sending calculating results,
and we will not keep the email address or leak it to others.
 Task ID
Task ID
 The online coding potential calculator will automatically assign an unique ID for each
calculating task. Users can use the Task ID to retrieve their calculating results.
For more details, please refer to our Quick Guide .
The online coding potential calculator will automatically assign an unique ID for each
calculating task. Users can use the Task ID to retrieve their calculating results.
For more details, please refer to our Quick Guide .
 NOTES: Task ID may be expired
after 7 days when it is created. The results of an expired task will removed from
the CPC server and can no longer be retrieved.
NOTES: Task ID may be expired
after 7 days when it is created. The results of an expired task will removed from
the CPC server and can no longer be retrieved.
 Hit Num
Hit Num
 How many blast hits are found for the input sequence.
How many blast hits are found for the input sequence.
 Hit Score
Hit Score
 For a true protein-coding transcript the hits are also likely to have higher
quality; i.e., the HSPs (High-scoring Segment Pairs) overall tend to have
lower E-value. Thus we define feature HIT SCORE as follows::
For a true protein-coding transcript the hits are also likely to have higher
quality; i.e., the HSPs (High-scoring Segment Pairs) overall tend to have
lower E-value. Thus we define feature HIT SCORE as follows::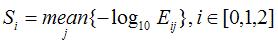
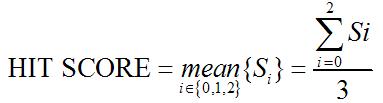
 Frame Score
Frame Score
 For a true protein-coding transcript most of the hits are likely to reside
within one frame, whereas for a true noncoding transcript, even if it matches
certain known protein sequence segments by chance, these chance hits are
likely to scatter in any of the three frames. Thus we define
feature FRAME SCORE to measure the distribution of the HSPs
among three reading frames:
For a true protein-coding transcript most of the hits are likely to reside
within one frame, whereas for a true noncoding transcript, even if it matches
certain known protein sequence segments by chance, these chance hits are
likely to scatter in any of the three frames. Thus we define
feature FRAME SCORE to measure the distribution of the HSPs
among three reading frames: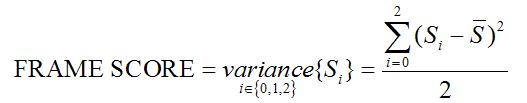
 FrameFinder ORF Coverage
FrameFinder ORF Coverage
 FrameFinder's orf coverage, A large COVERAGE OF THE PREDICTED ORF is
an indicator of good ORF quality (Slater, G.S.C. (2000) Algorithms
for the Analysis of Expressed Sequence Tags, University of Cambridge, Cambridge.) .
For more information, refer to
Pasteur FrameFinder Man Page
.
FrameFinder's orf coverage, A large COVERAGE OF THE PREDICTED ORF is
an indicator of good ORF quality (Slater, G.S.C. (2000) Algorithms
for the Analysis of Expressed Sequence Tags, University of Cambridge, Cambridge.) .
For more information, refer to
Pasteur FrameFinder Man Page
.
 FrameFinder LOG-ODDS SCORE
FrameFinder LOG-ODDS SCORE
 As suggested by the FrameFinder's author, the LOG-ODDS SCORE is an indicator
of the quality of a predicted ORF and the higher score, the higher the quality.
For more information, refer to
Pasteur FrameFinder Man Page
.
As suggested by the FrameFinder's author, the LOG-ODDS SCORE is an indicator
of the quality of a predicted ORF and the higher score, the higher the quality.
For more information, refer to
Pasteur FrameFinder Man Page
.
 FrameFinder ORF Type
FrameFinder ORF Type
 The ORF Type is the INTEGRITY OF THE PREDICTED ORF, that indicates whether an
ORF begins with a start codon and ends with an in-frame stop codon.
The ORF Type is the INTEGRITY OF THE PREDICTED ORF, that indicates whether an
ORF begins with a start codon and ends with an in-frame stop codon.
 Html View
Html View
 The html view of coding potential calculator online results.
The html view of coding potential calculator online results.
 UTRdb
UTRdb
 UTResource-DB (UTRdb) is a specialized database of 5' and 3' UnTRanslated sequences of eukaryotic
mRNAs cleaned and annotated based on RefSeq. For more detials, refer to
UTResource-DB Web Site.
UTResource-DB (UTRdb) is a specialized database of 5' and 3' UnTRanslated sequences of eukaryotic
mRNAs cleaned and annotated based on RefSeq. For more detials, refer to
UTResource-DB Web Site.
 RNAdb
RNAdb
 A comprehensive mammalian noncoding RNA database, includes >800 unique
experimentally studied ncRNAs, >1100 putative antisense ncRNA and almost 20000
putative ncRNAs identified in high-quality murine and human cDNA libraries.
For more detials, refer to RNAdb Web Site.
A comprehensive mammalian noncoding RNA database, includes >800 unique
experimentally studied ncRNAs, >1100 putative antisense ncRNA and almost 20000
putative ncRNAs identified in high-quality murine and human cDNA libraries.
For more detials, refer to RNAdb Web Site.
